Abstract
Rhizosphere is the hub for microbial activities where microbes and plants interact with complex signaling mechanisms. Plants release various metabolites in response to environmental factors which are significant in shaping rhizospheric microbial communities. These microbes develop symbiotic relation with plants by quorum sensing signals and regulate various microbial activities including biofilm formation. Biofilms are important in inhabiting rhizosphere and provide platform for cell-to-cell microbial interactions. Biofilm- forming rhizobacteria can successfully colonize plant roots and establish symbiotic relations with host. During this association, rhizobacteria are flourished by using plant root exudates, while the bacteria benefit the plants by synthesizing phytohormones, locking soil minerals for plant, protecting them from pathogenic invasions and enhancing plant immunity by improving plant tolerance against various environmental conditions. Indole is an effector molecule in regulating bacterial gene expression related to biofilm production. These interactions are coordinated by bacterially released phytohormones mainly auxin which act as key factor in regulating plant-microbe symbiotic interactions. It is characterized as inter- kingdom signaling molecule that coordinates various plant and rhizobacterial activities. Thus, understanding the nature and interacting behaviors of these molecules would lead to the exploitation of plant growth-promoting rhizobacteria for better plant growth in agricultural fields.
Keywords
- quorum sensing
- auxin
- biofilms
- rhizosphere
- bacterial signaling
- biofertilizers
- PGPR
1. Introduction
Rhizosphere is the most complex ecosystem where plants interact with billions of microbes that colonize plant roots. Symbiotic interactions between host and its associated microbiota are ecologically significant in maintaining ecosystem stability. The associated rhizobacteria is also referred to as the second genome as it allows the host metabolism to trigger various genomic and functional repertoire to deal with a wide range of ecological challenges [1]. Rhizosphere is a hotspot for various plant-rhizobacterial activities derived mainly through chemical signals. Plants can preferentially interact with tiny soil creatures recruiting them according to their own ecological demands. Plants can filter rhizospheric bacteria by secreting certain chemical messengers that disproportionally increase relative abundance of mutualistic rhizobacteria and decrease antagonistic species in rhizosphere [2, 3]. These chemical messengers can either be plant primary metabolites such as carbohydrates, organic acids, lipids, amino acids, nucleic acids, or secondary metabolites such as alkaloids, lignin, phenolics, terpenoids, sterols, steroids, essential oils, etc. These metabolites are released in the form of root exudates and are a source of nitrogen and carbon for rhizobacteria [4]. Crosstalk among microbes and pathogens is an important mechanism to ensure successful root colonization and development of symbiotic relationship with their photobiont partner. Generally, rhizospheric communications are identified as inter- and intra-species microbial signaling, plant to microbes signaling and vice versa [5]. Quorum sensing (QS) is the fundamental means of rhizobacterial crosstalk. These sensing signals rely on inter- and intra-species microbial information that coordinate and control bacterial cell-to-cell activities in microbial communities. QS directs various bacterial functions involving biofilm formation [6]. Some phytohormones released by rhizobacteria also act as signaling molecules. Majority of rhizobacteria, that is, 80% are able to produce auxins (IAA) by utilizing tryptophan precursor present in plant root exudates. IAA is an effector molecule in bacterial gene expression related to indole signaling molecules that regulate biofilm production [7]. These signals are the chief elements affecting the structure and physical heterogeneity of soil microbial communities which in turn influence plant resilience and plasticity. These plant-microbial cross communications are dynamic and interdependent on both partners as well as rhizospheric ecological parameters [8]. Microbial processes have great impact on ecological parameters by regulating various biogeochemical cycles to establish a compatible niche for healthier plants. Understanding these soil interactions occurring among microorganisms and their host is critical for the manipulation of untapped potential of microbiome and their usability in fields of environmental and sustainable agriculture [9]. Deciphering rhizospheric signals could enable scientists to exploit rhizobacteria to promote plant growth and production. The overall plant growth depends on the soil microbiota and its association with host plant [10]. Hence, rhizospheric engineering can provide a healthier environment for plant growth but rhizospheric engineering require understanding of these signals. The current chapter unentangles the complexity of rhizospheric signals among microbes and their photobionts and the significance of these signalings in the development of symbiotic associations.
2. Decrypting phyto-microbiome signals
As described earlier, three major categories of rhizospheric signals are plant-to-microbe signaling which is regulated by low molecular weight molecules secreted in the form of root exudates and are responsible for establishing plant-microbes interactions, inter- and intra-microbial signaling regulated by QS which is responsible for diversity and contemporizing microbial behavior, and lastly, microbe to plant signals regulated by microbially produced substances responsible for altering plant physiology and enhancing immunity to cope with environmental fluctuations [11]. Consequently, plants and microbes develop cooperative relationship which improves plant growth and health.
2.1 Microbial intra-kingdom crosstalk
Quorum sensing is bacterial signaling process which is initiated through detection of exogenous chemical signals. Bacterial cells synthesize and release chemical signals known as autoinducers (AIs) which in turn regulate the synthesis of various microbial functions such as biofilm formation, virulence, adhesion, motility, metabolism, and symbiotic association [12]. QS signals provide bacteria to survive competitive environment, increase microbial survival under various habitats and assist microbial rhizospheric colonization to develop plant microbial associations. Various quorum sensing signaling networks have been identified among rhizobacterial species such as diketopiperazines (DKPs), acylated homoserine lactones (AHLs), diffusible signaling factor (DSF), phytohormones, antibiotics and secondary metabolites, etc. [13, 14]. Quorum sensing signals are categorized as acyl-homoserine lactone (AHLs) which are commonly produced by Gram-negative bacteria and autoinducer peptides (AIPs) which are commonly produced by Gram-positive bacteria, out of AIPs autoinducer type 2 have properties of both AHLs and AIPs, hence, can be produced by both Gram-negative and Gram-positive bacteria. AHL-regulated QS signaling is known as the LuxI/Rlike QS system [15].
Rhizobacteria recognize specific signals within their vicinity and stimulate the synthesis of QS that control transcription of various microbial mechanisms [16]. Plant-beneficial bacteria also protect plants by releasing certain signals such as acylase and lactonase, which degrade pathogenic AHLs or release VOCs to compete with rhizospheric phytopathogens. Besides this, most of the rhizobacteria have ability to produce antimicrobial compounds such as cyclic-lipodepsipeptides and polycyclic tetramate macrolactams that hinder the phytopathogenic abundance and colonization [17]. A complex communal signaling network mechanism initiates as a result of interactions of microbial QS with plant signals. These signal exchanges occur both outside and inside plants [18].
2.2 Microbial inter-kingdom crosstalk
Among these QS, AHLs are commonly produced signaling molecules in rhizobacteria which also act as interkingdom signaling molecules as it also regulates expression of various genes in plants [19, 20]. Some AHLs such as cyclodipepetides and their derivatives also modulate auxin-responsive genes expression in plant roots [21]. Moreover, AHLs also regulate plant auxin to cytokinin ratio by triggering genes responsible for upregulation of auxin and downregulation of cytokinin [22]. AHLs, that is, N-butanoyl homoserine lactone and N-hexanoyl homoserine lactone, greatly impact plant physiology. Short-chain AHLs regulate plant growth especially root elongation [23]. QS also promotes signals for nodulations. These signals are responsible for regulating plant defense systems related to systemic acquired resistance (SAR) and induced systemic resistance (ISR) [24]. There is a molecular mechanism known as microbe-associated molecular patterns (MAMPs) that allows recognition based on highly conserved signature molecular patterns of microbes that are unique to the whole class. This is similar to pattern recognition receptors (PRRs) present in plants that recognize microbial compounds within root vicinity [25, 26]. Identification of unique MAMPs by phytoreceptors trigger signals from roots that modulate signaling pathways of various phytohormones such as salicylic acid, jasmonic acid, ethylene, etc. which trigger the expression of genes related to plant secondary metabolites and defensive proteins to activate resistance within plants [15]. Microbial QS interact with VOCs within rhizosphere. VOCs are significant for long-distance interactions with rhizobacteria and responsible for managing microbial populations and plant-microbe associations. VOCs serve as inter- and intra-species signaling molecules by regulating gene expression of various bacterial functions including biofilm formation. VOCs also modulate plant physiology, that is, root system architecture and plant hormonal signaling [27, 28]. Microbial VOCs trigger and interfere with auxin-signaling pathway in host plants [29]. Microbial diffusible signaling factor is also responsible for regulating plant immunity by trigging various plant defensive signaling. Similarly, various Gram-positive bacteria use peptides as QS to colonize plant roots which is also considered to regulate defensive signaling within plants [30, 31]. Moreover, QS also regulates nodulation in leguminous plants. Flavonoids present in root exudates enhance microbial gene expression for AHL synthesis [32]. Plants possess quorum quenching (QQ) mechanisms to QS signaling. Plants have adapted various mechanisms such as enzymatic degradation, biosynthesis and secretion of anti-QS compounds, disruption of binding, receptor, and regulatory sites [33, 34]. Sometimes, plants produce QS-intimate molecules, that is, p-coumaric acid, isothiocyanate sulforaphane, curcumin, patulin, etc. [35, 36, 37, 38]. Among these molecules, alkamides andN-acylethanolamines (NAEs) are excellent contenders regarding their chemistry and structure to act as AHL [38]. These AHL analogs interfere with pathogenic QS regulatory pathways inhibiting their gene expression [39].
2.3 Rhizospheric phyto-signaling
The chemicals released by plants directly influence the structure of microbial communities by influencing abundance and diversity of rhizobacteria. Root exudates are important for microbial root colonization. Metabolically active bacterial species are attracted toward root exudates [40]. Root exudation occurs through active and passive mechanisms. Secretion of low molecular weight signals is a passive process whereas other metabolites such as mucopolysaccharides are released actively. ATP-binding cassette transporters such as ABC transporters are involved in exudation phenomena [41]. The microbes within root vicinity use the substances present in root exudates. These substances act as plant pheromones that either stimulate or antagonize other microbes. Moreover, the composition of root exudates also modifies microbial dynamics which favors abundance of plant growth-promoting rhizobacteria (PGPR) and prevents pathogenic microbial growth [42]. Hence, root exudate mediates a tripartite signal among PGPR, pathogens, and the host plant [43]. Root exudates can either be low molecular weight compounds such as sugars, amino acids, organic acids, phenolics, and other secondary metabolites or high molecular weight compounds, that is, mucopolysaccharides [44]. The quantity and quality of exudates depend on various plant physiological parameters such as plant species, nutritional status, growth stage, etc. These compounds act as sources of nitrogen and carbon for microbes. The presence of these catabolic pathways is significant in root colonization and disease suppression [45]. Amino acids and carbohydrates act as microbial pheromones whereas protein molecules are essential for antagonistic signaling [46, 47]. For instance, lectin proteins released as root exudates assist in both symbiosis and defense responses [48]. Moreover, arabinogalactans (mucopolysaccharides) exuded from roots attracts PGPR and repel rhizospheric pathogens [49]. Besides amino acids, proteins, and carbohydrates, a considerable amount of phytohormones are also present in root exudates. Salicylic and jasmonic acids are involved in shaping rhizospheric microbial community. Microbial abundance is relatively high in the presence of these phytohormones and provoke plant immunity [50]. Although volatile organic compounds (VOCs) are quite significant aboveground signaling molecules, however, minute quantity of VOCs is also released in the form of root exudates. These compounds attract and kill root-colonizing rhizospheric pathogens [51].
Transcriptional profiling showed that expression of various microbial genes is regulated by root exudates. Phytochemicals direct various bacterial processes involved in rhizosphere colonization such as chemotaxis, motility, phase variation, membrane integrity, environmental resistance, and nutrient sequestration [45]. Root exudates also trigger microbial biofilm-related pathways. Biofilm formation is significant in rhizospheric colonization, adhesion to seeds, and roots [52]. Microbial motility in response to phytochemicals is crucial for recruiting rhizobacteria within plant vicinity [53]. Rhizospheric microbial activities determine the amount of root exudates which in turn affect patterns of root distribution toward rhizospheric nutrient pools. These exudates induce microbial chemotaxis by trigging the expression of various proteins related to methyl-accepting chemotaxis (MCP) such as Mcp (A, B, C), Tlp (A, B, C), YvaQ , YoaH, HemAT, and YfmS. These proteins adhere to the chemoreceptors when concentration of root exudated reaches to threshold. The binding of ligand to the ligand binding domain of chemoreceptors induces autophosphorylation of protein followed by transfer of phosphoryl group to response regulatory protein which interacts with flagellar motor and controls microbial movement responsible for chemotaxis [54, 55].
2.4 Auxins mediated cross-kingdom signal transmission
Auxins (IAA) are master phyto-hormone that control majority of plant functions. Besides plant, rhizobacteria also have ability to produce auxins as a result of their defensive mechanism. Tryptophan is most significant amino acid exuded by plants. Microbes in root vicinity detoxify tryptophan and produce auxins [56]. PIN proteins mediated auxin flux allows initiation of cell-to-cell communicating network [57]. Auxins act as signaling molecules for communication between microbes to coordinate their activities. Auxins also trigger diverse plant metabolic functions. Modification of root system architecture is one of the most promising features regulated by auxin producing PGPR [58]. Auxins also act as phytochemicals that regulate rhizobacterial chemotaxis. IAA signals received by bacterial methyl accepting proteins present in chemoreceptors initiate flagellar movement in rhizobacteria. Moreover, IAA also serves as nutrient reservoir in rhizosphere for microbes [59, 60]. IAA also enhances competence within rhizosphere. IAA-producing bacteria are more competent and successful in colonizing rhizosphere. This indicates that IAA might have some evolutionary significance that enhances bacterial survival under various environmental conditions. It is considered that IAA provides protection to rhizobacteria under biotic and abiotic stress conditions ensuring their survival [61]. IAA also interacts with QS molecules and regulates secretion of extracellular polymeric substances, lipopolysaccharides, and formation of biofilms [7]. Hence, rhizobacterial colonization is regulated by the interaction of IAA and QS signals and their high survival is also linked with biofilm production. IAA plays important role in modulating gene expression of bacterial cells and making them more competent [56]. Transcriptome analysis revealed that IAA signaling causes downregulation and upregulation of various respiration affecting genes in rhizobacteria [62].
Auxin signaling is significant in root morphological processes such as root hair formation and development of root tips. Plant phenotypic plasticity relases on auxin signaling and transport. It induces hyperpolarization that causes physicochemical alterations in plasma membrane [63]. Plant root morphological alterations depends on auxin concentration. Auxin primarily induces formation of primary and lateral roots under optimum concentrations and adventitious root formation under higher concentrations [64]. Moreover, auxins also initiate the nodulation process in leguminous plants. Flavonoids interact with auxin-signaling pathway and inhibit auxin biosynthesis and initiate the nodulation process. Auxin signaling controls various processes of nodulation, that is, formation of bacteroids, vascular bundles, number of nodules, etc. [65]. Auxins also regulates signaling pathways related to other phytohormones. It regulates transcription of 1-aminocyclopropane-l-carboxylic acid (ACC) that regulate ACC deaminase production which in the presence of low ethylene converts to α-ketobutyrate and ammonia. Hence, auxins interact with ethylene and reduce effects of environmental stress on plants [66, 67]. Coordination of auxins with cytokinins regulates cell division and differentiation at root meristem resulting in root growth. Auxins initiate formation of lateral roots while interactions of auxins with ethylene are significant for the development of root hair and elongation. Overall mechanism of root growth is regulated by auxin-derived phytohormonal coordination. Similarly, brassinosteroids, gibberellins, strigolactones act synergistically with auxins to regulate plant physiological processes [68, 69]. These underlying signal coordinations are responsive for plant root adjustments to external stimuli through inducing primary root elongation, lateral roots, and root hair formation [70, 71]. The overall mechanism of signaling crosstalk is illustrated in Figure 1.
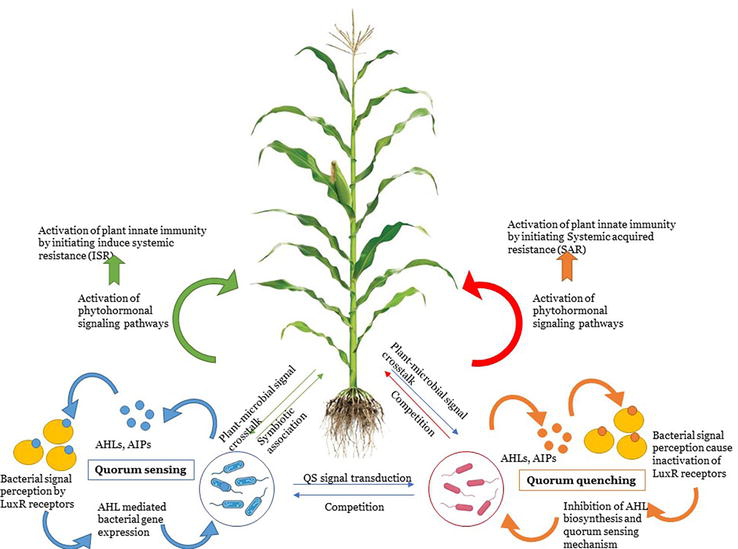
Figure 1.
Signal crosstalk among plant-Rhizobacteria and phytopathogens.
3. Signal-mediated phyto-microbial symbiotic responses
These signaling networks provide benefits to both partners in regulating symbiotic associations.
3.1 Rhizospheric colonization by biofilm formation
Microbial quorum sensing signals are involved in biofilm production that facilitates rhizobacteria to colonize plant roots and rhizosphere. Rhizobacteria have ability to produce multicellular communities embedded within extracellular polymeric substances regulated by QS. This phenomenon is regulated by complex signaling network. QS-mediated bacterial chemotaxis is important in regulating early stages of biofilms. QS such as cyclic dimeric guanosine 3′,5′-monophosphate (c-di-GMP), cyclic dimeric adenosine 3′,5′-monophosphate (c-di-AMP), and cyclic adenosine 3′,5′-monophosphate (cAMP) are the frontline signaling molecules in the regulation of biofilm development [72]. Biofilms are bacterial protecting shells under unpleasant environmental conditions to ensure their survivability and colonization in rhizosphere and to plant roots. Biofilms provide protection by secreting extracellular polymer. Extracellular polymer of biofilms is composed of exopolysaccharides (EPS), nucleic acids, proteins, and lipids which help in sequestration of toxic substances by elevating catabolic gene expression. Higher bacterial cell density within biofilms provides larger gene pool that facilitates adhesion and other metabolic activities [73]. This synchronization is controlled by QS mediated cell-to cell contact. The internal structures are interconnected for communication and transfer of nutrients. EPS primarily contains carbohydrates which provide foundation for the adhesion of various substances (proteins, lipids, nucleic acids) within biofilm microenvironment [74]. The release of QS depends on bacterial cell density. When population size reaches the threshold value QS initiates the formation of biofilms. In
3.2 Development of root nodules
Rhizobia-legume symbiotic interactions are widely known for their unique character, that is, nodule formation on plant roots in which rhizobia provide nitrogen to plants and in return get dicarboxylates. This process is initiated by the phytochemical signals, that is, flavonoid. Flavonoids interact with bacterial QS to activate nodulation genes (nod, noe, and nol) which then synthesize nodulation factor (Nod), that is, lipo-chitooligosaccharides (LCOs) [83]. These signals are received by receptors on roots epidermis to initiate nodulation process. Besides flavonoids, some other molecules present in root exudates such as vanillin, chalcones, aldonic acid, betaines, and jasmonate also activate nod genes. These signals are specific to the plant species to ensure accurate host-microbe relationship [84]. Flavonoid signals determine auxin transport patterns in plant roots. Alterations in auxin transport patterns are important in the nodulation mechanism. During initial stages of nodule organogenesis, auxin transport is inhibited and later during initiation and differentiation of nodular cells, auxin transport increases, and auxin accumulates in nodule primordium. This auxin influx and efflux is controlled by PIN and AUX1 genes present in plant roots. Flavonoids itself can regulate PIN activation and localization for auxin transport. Plant signals trigger reduction in auxin efflux in response to rhizobial encounter leading to auxin accumulation in pericycle and inner cortex. Moreover, exogenous auxin (bacterially produced) also disrupts plant phytohormonal homeostasis to adjust auxin at permissive levels for nodulation [85, 86]. Rhizobia produce auxin that enhances plant root growth, cell division, differentiation, and nodule formation. It also increased the number of nodules which is associated with biological nitrogen fixation ability [87]. During nodulation, auxins also interact with other phytohormones (such as abscisic acid) that inhibit lateral root formation and stimulate nodulation.
3.3 Root architecture system
The impact of rhizobacteria on plant root system architecture is significant in plant growth induction. PGPR modify plant root system architecture by their phytohormone production ability which interact with plant hormonal balance. An enhanced lateral root with more branching pattern is one of the more distinctive impacts of PGPRs on plants [88]. The overall root architecture is the dynamic response of roots toward soil localized nutrient resources through meristematic activity. Root architecture is linked with nutrient acquisition, thus, roots with higher surface area can interact more with soil particles, soil water, and microorganisms. PGPR-associated roots displayed increased rigidity with increased length and diameter of the roots as well as the cortex and aerenchyma space, increased protoxylem poles, and metaxylem vessel components with enhanced flavonoid and phenolic contents. PGPR releases phytohormones which manipulates plant hormonal homeostasis. Impact of IAA on the architecture and growth of roots is very much significant [89]. Auxin to cytokinin ratio is important in shaping plant root system. IAA regulates a wide range of processes involved in plant growth and development, low concentrations of IAA can promote primary root elongation, whereas high concentrations of IAA promote the growth of lateral roots, shorten primary root length, and promote root hair formation [90]. The concentration of auxin in the plants changes according to its uptake and excretion from tissues, its production from tryptophan, and the generation of IAA conjugates [91]. Auxins encourage root development to its full potential. Moreover, its increased concentrations reduce root growth [92].
3.4 Regulation of plant defense responses
PGPR-associated plants configure high tolerance to unfavorable environmental conditions. PGPR induces innate immune response in plants against phytopathogens and also PGPR regulates defense responses in plants by activating defense-related signaling pathways and prepares plants for accurate response. Plants recognize phytopathogens through pattern recognition receptors (PRRs) by recognizing pathogen-associated molecular patterns (PAMPs or MAMPs) to initiate signal cascades for the regulation of specific transcription factors to activate multiple intracellular defense responses [20]. This induction of signal cascade is either hormone dependent or independent. Rhizobacteria-produced phytohormones such as auxins, abscisic acid, gibberellins, and cytokinins also contribute toward these signal transductions. This stimulates intracellular defense responses [93]. AHLs and DSFs act as signaling molecule in plant-pathogenic interactions. This signal is perceived by plants and initiate the phytohormone-mediated biosynthesis of phytoanticipins or phytoalexins [94]. PGPRs can also recognize specific phytopathogens. PGPR-produced salicylic acid (SA) regulate plant defense response known as systemic acquired resistance (SAR). It binds with non-repressor of pathogenesis-related genes (NPR1). SA pathway is also involved in regulating flavonoid signaling which inhibits pathogenic proliferations [95]. SA-independent PGPR-mediated plant defense system is referred to as induced systemic resistance (ISR). This pathway includes jasmonic acid and ethylene signals. Invasion of PGPR trigger MAMPs recognition leading to the activation of ISR mechanism [95]. The development of symbiotic associations between plants and microbes has been illustrated in Figure 2.
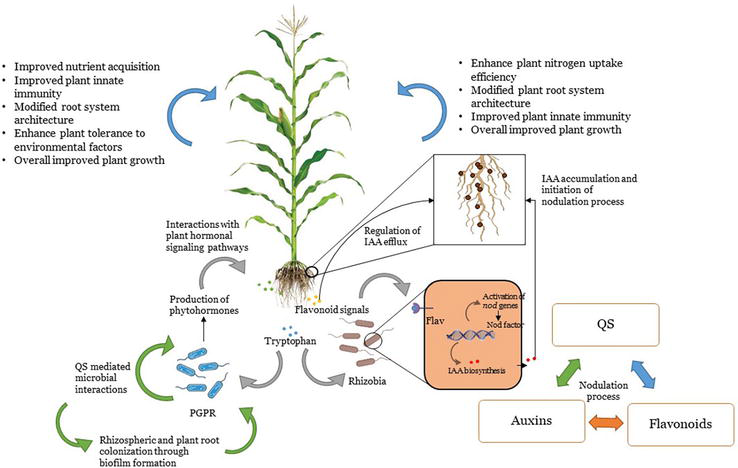
Figure 2.
Development of symbiotic associations between plants and beneficial bacteria by regulating plant phytohormonal pathways.
4. Agronomic efficiencies of symbiotic signaling
Plant microbial signaling mechanisms are responsible for the development of plant-microbial associations which is crucial for sustainable agricultural. Nutrient acquisition is a significant consequence of these associations. A cascade of signals regulates the continuous flow of nutrient exchange between both partners.
4.1 PGPR as biofertilizers
Various commercially available PGPR bioformulations are used as biofertilizers to increase crop productivity [96]. PGPR can restore the nutrient cycle between the soil, plant roots, and existing microorganisms [97]. Fertilizers containing nitrogen drive up manufacturing costs. Nitrogen biofixation is the first method of accelerating plant development which regulates nitrogenase activity and helps the plant regain its nitrogen equilibrium. In order to break the bond between the nitrogen atoms, the N2-fixing process carried out by microbes requires energy, which is supplied by readily available organic carbon. The root mutualistic bacteria may fix up to 30 kg/ha of pure nitrogen each year. The amount of nitrogen fixed by these bacteria can be useful for maintaining soil fertility over the long run. Biological N2-fixing PGPR is injected into crops and crop fields to promote growth, control disease, and maintain the nitrogen content in agricultural soil [98]. They are reliable, eco-friendly sources of the renewable nutrients needed to sustain the biology and health of the soil [99]. On the basis of their capacity to draw nutrients from the soil, fix atmospheric N2, promote the solubilization of nutrients, a variety of microbial taxa have been utilized commercially as effective biofertilizers [100].
4.2 PGPR as biocontrol agent
The utilization of these rhizobacteria by farmers in the field is currently insufficient. To address variability in the efficacy of the biocontrol system, combined activity of several bacterial strains is necessary. These rhizobacteria mixes can be employed as a seed treatment, which might help to cut down on the amount of bacterial inoculum needed. This will promote both the hostile activity of the bacteria on the root surface during the early root infection by the pathogens and their systemic dissemination over the surface of the growing root system [101].
4.3 PGPR in phytoremediation
Use of PGPR biofilms in phytoremediation technologies plays a significant role. PGPR enhances the qualities of plants employed in phytoremediation, such as biomass production, low-level pollutant absorption, plant nutrition, and health, however, it is crucial to pick PGPR that can last and be effective in phytoremediation procedures. Metal-tolerant PGPR hinders direct uptake of heavy metals to plants by converting them to less toxic forms as a result plant growth is improved [102]. When PGPR are applied to a polluted site, they increase the capacity of the plants that grow there to detoxify synthetic chemicals and preserve soil structure. Additionally, PGPR reduces the toxicity of heavy metals by changing their bioavailability in plants [103].
5. Conclusion and future prospects
Microbial interactions with plants are carried out through an interconnected signaling network that work inter- and intra-specifically. These signaling mechanisms decipher the microbiome communications related to plant productivity. Various plant metabolites released through roots in the form of root exudates attract rhizobacteria that interact by producing quorum-sensing signals. These signals are responsible for various bacterial activities in rhizosphere. Digging into their molecular pattern is necessary to take their full advantage. Manipulating these signaling pathways can help in rhizospheric engineering which will be beneficial for sustainable agriculture. Expanding research related to metagenomic analysis of rhizomicrobiome is important to understand whole infrastructure of symbiotic associations. Moreover, use of appropriate PGPR and their commercialization is necessary as they are more economic and environmentally safe compared to chemical fertilizers. Thereby, utilization of PGPR base biofertilizers are much appreciated.
References
- 1.
Klein M, Stewart JD, Porter SS, Weedon JT, Kiers ET. Evolution of manipulative microbial behaviors in the rhizosphere. Evolutionary Applications. 2022; 15 (10):1521-1536. DOI: 10.1111/eva.13333 - 2.
Bakker PA, Berendsen RL, Van Pelt JA, Vismans G, Yu K, Li E, et al. The soil-borne identity and microbiome-assisted agriculture: Looking back to the future. Molecular Plant. 2020; 13 (10):1394-1401. DOI: 10.1016/j.molp.2020.09.017 - 3.
Li E, de Jonge R, Liu C, Jiang H, Friman VP, Pieterse CM, et al. Rapid evolution of bacterial mutualism in the plant rhizosphere. Nature Communications. 2021; 12 (1):1-3. DOI: 10.1038/s41467-021-24005-y - 4.
Olanrewaju OS, Ayangbenro AS, Glick BR, Babalola OO. Plant health: Feedback effect of root exudates-rhizobiome interactions. Applied Microbiology and Biotechnology. 2019; 103 (3):1155-1166. DOI: 10.1007/s00253-018-9556-6 - 5.
Venturi V, Keel C. Signaling in the rhizosphere. Trends in Plant Science. 2016; 21 (3):187-198. DOI: 10.1111/1462-2920.13571 - 6.
Zhou L, Zhang Y, Ge Y, Zhu X, Pan J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Frontiers in Microbiology. 2020; 11 :589640. DOI: 10.3389/fmicb.2020.589640 - 7.
Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harbor Perspectives in Biology. 2011; 3 (4):1-13. DOI: 10.1101/cshperspect.a001438 - 8.
Hassan MK, McInroy JA, Kloepper JW. The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: A review. Agriculture. 2019; 9 (7):142. DOI: 0.3390/agriculture9070142 - 9.
Hakim S, Naqqash T, Nawaz MS, Laraib I, Siddique MJ, Zia R, et al. Rhizosphere engineering with plant growth-promoting microorganisms for agriculture and ecological sustainability. Frontiers in Sustainable Food Systems. 2021; 5 :617157. DOI: 10.3389/fsufs.2021.617157 - 10.
Masood S, Zhao XQ , Shen RF. Bacillus pumilus promotes the growth and nitrogen uptake of tomato plants under nitrogen fertilization. Scientia Horticulturae. 2020;272 :109581. DOI: 10.1016/j.scienta.2020.109581 - 11.
Altaf M, Khan M, Ahmad S, Abulreesh HH, Ahmad I. Quorum sensing in plant growth-promoting rhizobacteria and its impact on plant-microbe interaction. In: Dhananjaya Pratap Singh DP, Singh HB, Prabha P, editors. Plant-Microbe Interactions in AgroEcological Perspectives. Singapore: Springer; 2017. pp. 311-331. DOI: 10.1007/978-981-10-5813-4_16 - 12.
Checcucci A, Marchetti M. The rhizosphere talk show: The rhizobia on stage. Frontiers in Agronomy. 2020; 2 :591494. DOI: 10.3389/fagro.2020.59149 - 13.
Cortez M, Handy D, Headlee A, Montanez C, Pryor S, Cutshaw K, et al. Quorum sensing in the rhizosphere. In: Horwitz BA, Mukherjee PK, editors. Microbial Cross-Talk in the Rhizosphere. Singapore: Springer; 2022. pp. 99-134. DOI: 10.1007/978-981-16-9507-0_5 - 14.
Mondal S, Baksi S. Signalling of rhizosphere microbiomes: Benign and malign borders. In: Arora NK, Bouizgarne B, editors. Microbial BioTechnology for Sustainable Agriculture. Vol. 1. Singapore: Springer; 2022. pp. 237-260. DOI: 10.1007/ 978-981-16-4843-4_7 - 15.
Jamil F, Mukhtar H, Fouillaud M, Dufossé L. Rhizosphere signaling: Insights into plant–rhizomicrobiome interactions for sustainable agronomy. Microorganisms. 2022; 10 (5):899. DOI: 10.3390/microorganisms10050899 - 16.
Bramhachari PV. Implication of quorum sensing system in biofilm formation and virulence. Springer; Singapore. 2019. p. 28. DOI: 10.1007/978-981-13-2429-1 - 17.
Brescia F, Marchetti-Deschmann M, Musetti R, Perazzolli M, Pertot I, Puopolo G. The rhizosphere signature on the cell motility, biofilm formation and secondary metabolite production of a plant-associated Lysobacter strain. Microbiological Research. 2020; 234 :126424. DOI: 10.1016/j.micres.2020.126424 - 18.
van der Burgh AM, Joosten MH. Plant immunity: Thinking outside and inside the box. Trends in Plant Science. 2019; 24 (7):587-601. DOI: 10.1016/j.tplants.2019.04.009 - 19.
Singh A, Chauhan PS. N-acyl homoserine lactone mediated quorum sensing exhibiting plant growth-promoting and abiotic stress tolerant bacteria demonstrates drought stress amelioration. Journal of Pure and Applied Microbiology. 2022; 1 :669-684. DOI: 10.22207/JPAM.16.1.69 - 20.
Bukhat S, Imran A, Javaid S, Shahid M, Majeed A, Naqqash T. Communication of plants with microbial world: Exploring the regulatory networks for PGPR mediated defense signaling. Microbiological Research. 2020; 238 :126486. DOI: 10.1016/j.micres.2020.126486 - 21.
Ortiz-Castro R, Campos-García J, López-Bucio J. Pseudomonas putida andPseudomonas fluorescens influence Arabidopsis root system architecture through an auxin response mediated by bioactive cyclodipeptides. Journal of Plant Growth Regulation. 2020;39 (1):254-265. DOI: 10.1007/s00344-019-09979-w - 22.
Nadarajah K, Abdul Rahman NS. Plant–microbe interaction: Aboveground to belowground, from the good to the bad. International Journal of Molecular Sciences. 2021; 22 (19):10388. DOI: 10.3390/ijms221910388 - 23.
Phour M, Sehrawat A, Sindhu SS, Glick BR. Interkingdom signaling in plant-rhizomicrobiome interactions for sustainable agriculture. Microbiological Research. 2020; 241 :126589. DOI: 10.1016/j.micres.2020.126589 - 24.
Shrestha A, Grimm M, Ojiro I, Krumwiede J, Schikora A. Impact of quorum sensing molecules on plant growth and immune system. Frontiers in Microbiology. 2020; 11 :1545. DOI: 10.3389/fmicb.2020.01545 - 25.
Pantigoso HA, Newberger D, Vivanco JM. The rhizosphere microbiome: Plant–microbial interactions for resource acquisition. Journal of Applied Microbiology. 2022; 133 (5):2864-2876. DOI: 10.1111/jam.15686 - 26.
Yu J, Tu X, Huang AC. Functions and biosynthesis of plant signaling metabolites mediating plant–microbe interactions. Natural Product Reports. 2022; 39 (7):1393-1422. DOI: 10.1039/D2NP00010E - 27.
Poveda J. Beneficial effects of microbial volatile organic compounds (MVOCs) in plants. Applied Soil Ecology. 2021; 168 :104118. DOI: 10.1016/j.apsoil.2021.104118 - 28.
Weisskopf L, Schulz S, Garbeva P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nature Reviews. Microbiology. 2021; 19 (6):391-404. DOI: 10.1038/s41579-020-00508-1 - 29.
Tyagi S, Mulla SI, Lee KJ, Chae JC, Shukla P. VOCs-mediated hormonal signaling and crosstalk with plant growth promoting microbes. Critical Reviews in Biotechnology. 2018; 38 (8):1277-1296. DOI: 10.1080/07388551.2018.1472551 - 30.
Monnet V, Juillard V, Gardan R. Peptide conversations in Gram-positive bacteria. Critical Reviews in Microbiology. 2016; 42 (3):339-351. DOI: 10.3109/1040841X.2014.948804 - 31.
He YW, Deng Y, Miao Y, Chatterjee S, Tran TM, Tian J, et al. DSF-family quorum sensing signal-mediated intraspecies, interspecies, and inter-kingdom communication. Trends in Microbiology. 2022; 5 :36-50. DOI: 10.1016/j.tim.2022.07.006 - 32.
Babenko LM, Romanenko КО, Iungin OS, Kosakovska IV. Acyl-homoserine lactones for crop production and stress tolerance of agricultural plants. Agricultural Biology [Sel'skokhozyaistvennaya Biologiya]. 2021; 56 (1):3-19. DOI: 10.15389/agrobiology.2021.1.3eng - 33.
Deryabin D, Galadzhieva A, Kosyan D, Duskaev G. Plant-derived inhibitors of AHL-mediated quorum sensing in bacteria: Modes of action. International Journal of Molecular Sciences. 2019; 20 (22):5588. DOI: 10.3390/ijms20225588 - 34.
Billot R, Plener L, Jacquet P, Elias M, Chabrière E, Daudé D. Engineering acyl-homoserine lactone-interfering enzymes toward bacterial control. Journal of Biological Chemistry. 2020; 295 (37):12993-13007. DOI: 10.1074/jbc.REV120.013531 - 35.
Jakobsen TH, van Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M, et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrobial Agents and Chemotherapy. 2012; 56 (5):2314-2325. DOI: 10.1128/AAC.05919-11 - 36.
Ganin H, Rayo J, Amara N, Levy N, Krief P, Meijler MM. Sulforaphane and erucin, natural isothiocyanates from broccoli, inhibit bacterial quorum sensing. Medicinal Chemistry Communications. 2013; 4 (1):175-179. DOI: 10.1039/C2MD20196H - 37.
Hartmann A, Rothballer M, Hense BA, Schröder P. Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Frontiers in Plant Science. 2014; 5 :131. DOI: 10.3389/fpls.2014.00131 - 38.
Aziz M, Chapman KD. Fatty acid amide hydrolases: An expanded capacity for chemical communication? Trends in Plant Science. 2020; 25 (3):236-249. DOI: 10.1016/j.tplants.2019.11.002 - 39.
Kamath A, Shukla A, Patel D. Quorum sensing and quorum quenching: Two sides of the same coin. Physiological and Molecular Plant Pathology. 2022; 123 :101927. DOI: 10.1016/j.pmpp.2022.101927 - 40.
Wang NQ , Kong CH, Wang P, Meiners SJ. Root exudate signals in plant–plant interactions. Plant, Cell & Environment. 2021; 44 (4):1044-1058. DOI: 10.1111/pce.13892 - 41.
Vives-Peris V, de Ollas C, Gómez-Cadenas A, Pérez-Clemente RM. Root exudates: From plant to rhizosphere and beyond. Plant Cell Reports. 2020; 39 (1):3-17. DOI: 10.1007/s00299-019-02447-5 - 42.
Yu Y, Gui Y, Li Z, Jiang C, Guo J, Niu D. Induced systemic resistance for improving plant immunity by beneficial microbes. Plants. 2022; 11 (3):386. DOI: 10.3390/plants11030386 - 43.
Liu Y, Zhang N, Qiu M, Feng H, Vivanco JM, Shen Q , et al. Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection. FEMS Microbiology Letters. 2014;353 (1):49-56. DOI: 10.1111/1574-6968.12406 - 44.
Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nature Microbiology. 2018; 3 (4):470-480. DOI: 10.1038/s41564-018-0129-3 - 45.
Mavrodi OV, McWilliams JR, Peter JO, Berim A, Hassan KA, Elbourne LD, et al. Root exudates alter the expression of diverse metabolic, transport, regulatory, and stress response genes in rhizosphere Pseudomonas . Frontiers in Microbiology. 2021;12 :698. DOI: 10.3389/fmicb.2021.651282 - 46.
More SS, Shinde SE, Kasture MC. Root exudates a key factor for soil and plant: An overview. The Pharma Innovation Journal. 2020; 8 :449-459 - 47.
Pascale A, Proietti S, Pantelides IS, Stringlis IA. Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Frontiers in Plant Science. 2020; 10 :1741. DOI: 10.3389/fpls.2019.01741 - 48.
Reyes-Montaño EA, Vega-Castro NA. Plant lectins with insecticidal and insect static activities. In: Begum G, editors. Insecticides Agriculture and Toxicology. London, UK: IntechOpen; 2018. pp. 17-42. DOI: 10.5772/intechopen.74962 - 49.
Villa-Rivera MG, Cano-Camacho H, López-Romero E, Zavala-Páramo MG. The role of arabinogalactan type II degradation in plant-microbe interactions. Frontiers in Microbiology. 2021; 12 :1-14. DOI: 10.3389/fmicb.2021.730543 - 50.
Singh K, Chandra R, Purchase D. Unraveling the secrets of rhizobacteria signaling in rhizosphere. Rhizosphere. 2022; 21 :100484. DOI: 10.1016/j.rhisph.2022.100484 - 51.
Gfeller V, Huber M, Förster C, Huang W, Köllner TG, Erb M. Root volatiles in plant–plant interactions I: High root sesquiterpene release is associated with increased germination and growth of plant neighbours. Plant, Cell & Environment. 2019; 42 (6):1950-1963. DOI: 10.1111/pce.13532 - 52.
Zboralski A, Filion M. Genetic factors involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. Computational and Structural Biotechnology Journal. 2020;18 :3539-3554. DOI: 10.1016/j.csbj.2020.11.025 - 53.
Feng H, Fu R, Hou X, Lv Y, Zhang N, Liu Y, et al. Chemotaxis of beneficial rhizobacteria to root exudates: The first step towards root–microbe rhizosphere interactions. International Journal of Molecular Sciences. 2021; 22 (13):6655. DOI: 10.3390/ijms22136655 - 54.
López-Farfán D, Reyes-Darias JA, Matilla MA, Krell T. Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Frontiers in Microbiology. 2019;10 :78. DOI: 10.3389/fmicb.2019.00078 - 55.
Colin R, Ni B, Laganenka L, Sourjik V. Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiology Reviews. 2021; 45 (6):fuab038. DOI: 10.1093/femsre/fuab038 - 56.
Tariq A, Ahmed A. Auxins interkingdom signaling molecules. In: Hano C, editors. Plant Hormones: Recent Advances, New Perspectives and Applications. Vol. 25. London, UK: IntechOpen; 2022. p. 3. Doi: 10.5772/intechopen.95608 - 57.
Saritha M, Kumar P, Panwar NR, Burman U. Intelligent plant–microbe interactions. Archives of Agronomy and Soil Science. 2022; 68 (7):1002-1018. DOI: 10.1080/03650340.2020.1870677 - 58.
Ahmed A, Hasnain S. Auxins as one of the factors of plant growth improvement by plant growth promoting rhizobacteria. Polish Journal of Microbiology. 2014; 63 (3):261 - 59.
Duca D, Lorv J, Patten CL, Rose D, Glick BR. Indole-3-acetic acid in plant–microbe interactions. Antonie Van Leeuwenhoek. 2014; 106 (1):85-125. DOI: 10.1007/s10482-013-0095-y - 60.
Duca DR, Glick BR. Indole-3-acetic acid biosynthesis and its regulation in plant-associated bacteria. Applied Microbiology and Biotechnology. 2020; 104 (20):8607-8619. DOI: 10.1007/s00253-020-10869-5 - 61.
Zhang Y, Li Y, Hassan MJ, Li Z, Peng Y. Indole-3-acetic acid improves drought tolerance of white clover via activating auxin, abscisic acid and jasmonic acid related genes and inhibiting senescence genes. BMC Plant Biology. 2020; 20 (1):1-2. DOI: 10.1186/s12870-020-02354-y - 62.
Kunkel BN, Johnson JM. Auxin plays multiple roles during plant–pathogen interactions. Cold Spring Harbor Perspectives in Biology. 2021; 13 (9):a040022. DOI: 10.1101/cshperspect.a040022 - 63.
Napier R. The story of auxin-binding protein 1 (ABP1). Cold Spring Harbor Perspectives in Biology. 2021; 13 (12):a039909. DOI: 10.1101/cshperspect.a039909 - 64.
Ahmed A, Tariq A, Habib S. Interactive biology of auxins and phenolics in plant environment. In: Lone R, Shuab R, Kamili AN, editors. Plant Phenolics in Sustainable Agriculture. Singapore: Springer; 2020. pp. 117-133. DOI: 10.1007/978-981-15-4890-1_5 - 65.
Grover M, Bodhankar S, Sharma A, Sharma P, Singh J, Nain L. PGPR mediated alterations in root traits: Way toward sustainable crop production. Frontiers in Sustainable Food Systems. 2021; 4 :618230. DOI: 10.3389/fsufs.2020.61823 - 66.
Zemlyanskaya EV, Omelyanchuk NA, Ubogoeva EV, Mironova VV. Deciphering auxin-ethylene crosstalk at a systems level. International Journal of Molecular Sciences. 2018; 19 (12):4060. DOI: 10.3390/ijms19124060 - 67.
Bhardwaj S, Sharma D, Jan S, Singh R, Bhardwaj R, Kapoor D. Crosstalk of ethylene and other phytohormones in the regulation of plant development. Ethylene in Plant Biology. 2022; 23 :17-31. DOI: 10.1002/9781119744719.ch2 - 68.
Vissenberg K, Claeijs N, Balcerowicz D, Schoenaers S. Hormonal regulation of root hair growth and responses to the environment in Arabidopsis. Journal of Experimental Botany. 2020; 71 (8):2412-2427. DOI: 10.1093/jxb/eraa048 - 69.
Mazzoni-Putman SM, Brumos J, Zhao C, Alonso JM, Stepanova AN. Auxin interactions with other hormones in plant development. Cold Spring Harbor Perspectives in Biology. 2021; 13 (10):a039990. DOI: 10.1101/cshperspect.a039990 - 70.
Eichmann R, Richards L, Schäfer P. Hormones as go-betweens in plant microbiome assembly. The Plant Journal. 2021; 105 (2):518-541. DOI: 10.1111/tpj.15135 - 71.
Nakano M, Omae N, Tsuda K. Inter-organismal phytohormone networks in plant-microbe interactions. Current Opinion in Plant Biology. 2022; 68 :102258. DOI: 10.1016/j.pbi.2022.102258 - 72.
Mahto KU, Kumari S, Das S. Unraveling the complex regulatory networks in biofilm formation in bacteria and relevance of biofilms in environmental remediation. Critical Reviews in Biochemistry and Molecular Biology. 2022; 57 (3):305-332. DOI: 10.1080/10409238.2021.2015747 - 73.
Arnaouteli S, Bamford NC, Stanley-Wall NR, Kovács ÁT. Bacillus subtilis biofilm formation and social interactions. Nature Reviews. Microbiology. 2021; 19 (9):600-614. DOI: 10.1038/s41579-021-00540-9 - 74.
Wang Y, Bian Z, Wang Y. Biofilm formation and inhibition mediated by bacterial quorum sensing. Applied Microbiology and Biotechnology. 2022; 12 :1-7. DOI: 10.1007/s00253-022-12150-3 - 75.
Oh MH, Han K. AbaR is a LuxR type regulator essential for motility and the formation of biofilm and pellicle in Acinetobacter baumannii . Genes & Genomics. 2020;42 (11):1339-1346. DOI: 10.1007/s13258-020-01005-8 - 76.
Thi MT, Wibowo D, Rehm BH. Pseudomonas aeruginosa biofilms. International Journal of Molecular Sciences. 2020;21 (22):8671. DOI: 10.3390/ijms21228671 - 77.
Passos da Silva D, Matwichuk ML, Townsend DO, Reichhardt C, Lamba D, Wozniak DJ, et al. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nature Communications. 2019;10 (1):1-11. DOI: 10.1038/s41467-019-10201-4 - 78.
Jiang Q , Chen J, Yang C, Yin Y, Yao K. Quorum sensing: A prospective therapeutic target for bacterial diseases. BioMed Research International. 2019; 2019 :1-15. DOI: 10.1155/2019/2015978 - 79.
Li Y, Feng T, Wang Y. The role of bacterial signaling networks in antibiotics response and resistance regulation. Marine Life Science & Technology. 2022; 28 :1-6. DOI: 10.1007/s42995-022-00126-1 - 80.
Zarkan A, Liu J, Matuszewska M, Gaimster H, Summers DK. Local and universal action: The paradoxes of indole signalling in bacteria. Trends in Microbiology. 2020; 28 (7):566-577. DOI: 10.1016/j.tim.2020.02.007 - 81.
Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiology Reviews. 2010; 34 (4):426-444. DOI: 10.1111/j.1574-6976.2009.00204.x - 82.
Cui B, Chen X, Guo Q , Song S, Wang M, Liu J, et al. The cell–cell communication signal indole controls the physiology and interspecies communication of Acinetobacter baumannii . Microbiology Spectrum. 2022;10 (4):691-696. DOI: 10.1111/1758-2229.13100 - 83.
Acosta-Jurado S, Fuentes-Romero F, Ruiz-Sainz JE, Janczarek M, Vinardell JM. Rhizobial exopolysaccharides: Genetic regulation of their synthesis and relevance in symbiosis with legumes. International Journal of Molecular Sciences. 2021; 22 (12):6233. DOI: 10.3390/ijms22126233 - 84.
Smith DL, Gravel V, Yergeau E. Signaling in the Phytomicrobiome. Frontiers in Plant Science. 2017; 8 :611. DOI: 10.3389/fpls.2017.00611 - 85.
Zhang P, Jin T, Kumar Sahu S, Xu J, Shi Q , et al. The distribution of tryptophan-dependent indole-3-acetic acid synthesis pathways in bacteria unraveled by large-scale genomic analysis. Molecules. 2019; 24 (7):1411. DOI: 10.3390/molecules24071411 - 86.
Bosse MA, da Silva MB, de Oliveira NG, de Araujo MA, Rodrigues C, de Azevedo JP, et al. Physiological impact of flavonoids on nodulation and ureide metabolism in legume plants. Plant Physiology and Biochemistry. 2021; 166 :512-521. DOI: 10.1016/j.plaphy.2021.06.007 - 87.
Kohlen W, Ng JL, Deinum EE, Mathesius U. Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. Journal of Experimental Botany. 2018; 69 (2):229-244. DOI: 10.1093/jxb/erx308 - 88.
Erturk Y, Ercisli S, Haznedar A, Cakmakci R. Effects of plant growth promoting rhizobacteria (PGPR) on rooting and root growth of kiwifruit ( Actinidia deliciosa ) stem cuttings. Biological Research. 2010;43 (1):91-98. DOI: 10.4067/S0716-97602010000100011 - 89.
Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moënne-Loccoz Y, Muller D, et al. Plant growth-promoting rhizobacteria and root system functioning. Frontiers in Plant Science. 2013; 4 :356. DOI: 10.3389/fpls.2013.00356 - 90.
Remans R, Beebe S, Blair M, Manrique G, Tovar E, Rao I, et al. Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean ( Phaseolus vulgaris L.). Plant and Soil. 2008;302 (1):149-161. DOI: 10.1007/s11104-007-9462-7 - 91.
Lima JV, Tinôco RS, Olivares FL, Chia GS, de Melo Júnior JA, da Silva GB. Rhizobacteria modify root architecture and improve nutrient uptake in oil palm seedlings despite reduced fertilizer. Rhizosphere. 2021; 19 :100420. DOI: 10.1016/j.rhisph.2021.100420 - 92.
Facella P, Daddiego L, Giuliano G, Perrotta G. Gibberellin and auxin influence the diurnal transcription pattern of photoreceptor genes via CRY1a in tomato. PLoS One. 2012; 7 (1):e30121 - 93.
Vannier N, Agler M, Hacquard S. Microbiota-mediated disease resistance in plants. PLoS Pathogens. 2019; 15 (6):e1007740 - 94.
Nishad R, Ahmed T, Rahman VJ, Kareem A. Modulation of plant defense system in response to microbial interactions. Frontiers in Microbiology. 2020; 11 :1298. DOI: 10.3389/fmicb.2020.01298 - 95.
Pršić J, Ongena M. Elicitors of plant immunity triggered by beneficial bacteria. Frontiers in Plant Science. 2020; 11 :594530. DOI: 10.3389/fpls.2020.594530 - 96.
García-Fraile P, Menéndez E, Celador-Lera L, Díez-Méndez A, Jiménez-Gómez A, Marcos-García M, et al. Bacterial probiotics: A truly green revolution. In: Kumar V, Kumar M, Sharma S, Prasad R, editors. Probiotics and Plant Health. Singapore: Springer; 2017. pp. 131-162. DOI: 10.1007/978-981-10- 3473-2_6 - 97.
Martínez-Viveros O, Jorquera MA, Crowley DE, Gajardo GM, Mora ML. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. Journal of Soil Science and Plant Nutrition. 2010; 10 (3):293-319. DOI: 10.4067/S0718-95162010000100006 - 98.
Damam M, Kaloori K, Gaddam B, Kausar R. Plant growth promoting substances (phytohormones) produced by rhizobacterial strains isolated from the rhizosphere of medicinal plants. International Journal of Pharmaceutical Sciences Review and Research. 2016; 37 (1):130-136 - 99.
Sun B, Bai Z, Bao L, Xue L, Zhang S, Wei Y, et al. Bacillus subtilis biofertilizer mitigating agricultural ammonia emission and shifting soil nitrogen cycling microbiomes. Environment International. 2020;144 :105989. DOI: 10.1016/j.envint.2020.105989 - 100.
Schütz L, Gattinger A, Meier M, Müller A, Boller T, Mäder P, et al. Improving crop yield and nutrient use efficiency via biofertilization—A global meta-analysis. Frontiers in Plant Science. 2018; 8 :2204. DOI: 10.3389/fpls.2017.02204 - 101.
Akhtar M, Siddiqui ZA. Role of plant growth promoting rhizobacteria in biocontrol of plant diseases and sustainable agriculture. In: Maheshwari DK, editors. Plant Growth and Health Promoting Bacteria. Berlin, Heidelberg: Springer; 2010. pp. 157-195. DOI: 10.1007/978-3-642-13612-2_7 - 102.
Fatima H, Ahmed A. Indole-3-acetic acid synthesizing chromium-resistant bacteria can mitigate chromium toxicity in Helianthus annuus L. Plant, Soil and Environment. 2020;66 (5):216-221. DOI: 10.17221/581/2019-PSE - 103.
Habib S, Ahmed A. Comparative analysis of pre-germination and post-germination inoculation treatments of Zea mays L. to mitigate chromium toxicity in Cr-contaminated soils. Polish Journal of Environmental Studies. 2018;28 (2):597-607. DOI: 10.15244/pjoes/81570